Genetic Perturbation Predictive Modeling
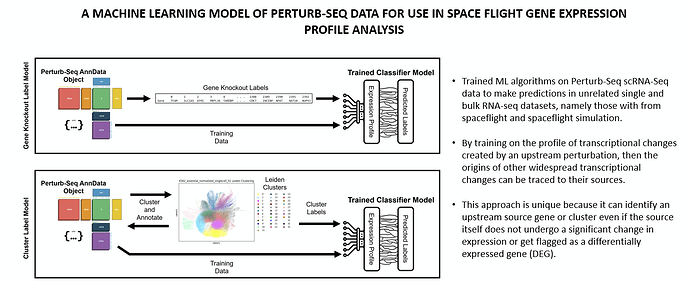
We are excited to introduce a new subgroup within the AI/ML AWG centered around predictive modeling using omic data created from genetic perturbation. We already have an existing project the involves training ML algorithms on Perturb-Seq scRNA-Seq data to make predictions in unrelated single and bulk RNA-seq datasets, namely those with from spaceflight and spaceflight simulation. The premise is that, by training on the profile of transcriptional changes created by an upstream perturbation, then the origins of other widespread transcriptional changes (such as those which occur in humans during spaceflight) can be traced to their sources. This approach is unique because it can identify an upstream source gene or cluster even if the source itself does not undergo a significant change in expression. This project already has a manuscript on biorxiv, but needs to be submitted to a journal, possibly BMC Bioinformatics, and will likely require some revision and expansion. Would like to see this published. From there we can explore avenues to expand upon the existing paradigm with new datasets and algorithms. There are several possible directions for this already on paper, but we are interested in any new avenues or ideas you might bring to the table.
Active Project Pre-Print:
https://doi.org/10.1101/2024.11.28.625741
Interest Response Form:
We are looking for members to fill the following roles/areas of attention:
Computation – Data processing and model training
Output Analysis – Gene set enrichment analysis and literature validation.
Research – Searching for new datasets, algorithms to apply, and studies to validate predictions against. Involves extensive literature review.
Code Organization – Maintaining and synchronizing versions, GitHub and Hugging Face maintenance. Familiarity with google collab notebooks would be helpful as we need to pivot towards that system.
Submission Experience – Expertise in navigating journal submissions.
Resources we are looking for:
Compute – The existing models were created using a limited dataset that fit within 164 gigs of memory, but we will need a larger server configured for remote access via collab. We have a 96 gig server which may be available for this as a last resort, but we would like to find a better solution.
OSDR human spaceflight RNA seq datasets - The existing project was created with the recently compiled human datasets in mind, but has never been used on them due to access limitations.